
DUMBOdqSPC5sqbsw: 2D big F1 spectral width SPC5 2Q/1Q correlation with DUMBO decoupling pulse program (TopSpin2.1)

DUMBOdqSPC5sqbsw: 2D big F1 spectral width SPC5 2Q/1Q correlation with DUMBO decoupling pulse program (TopSpin2.1)
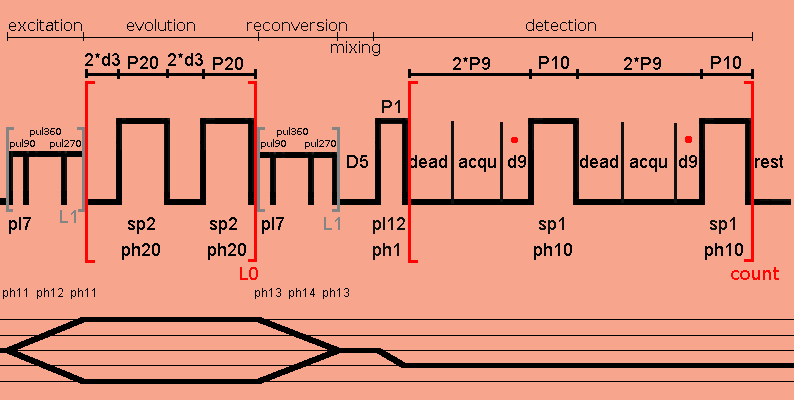
Since non-phase cycling is applied to the SPC5 excitation pulse, four-phase cycling is applied to the detection pulse P1 for selecting the 0Q -> -1Q coherence order jump, and four-phase cycling is applied to the SPC5 reconversion pulse for filtering DQ coherences.
;DUMBOdqSPC5sqbsw ;2D DQ-SQ proton-proton shift correlation ;with SPC5 DQ excitation/reconversion ;with homonuclear DUMBO decoupling DQ evolution without prepulses during t1 ;and windowed DUMBO acquisition ;S. P. Brown, A. Lesage, B. Elena and L. Emsley, J. Am. Chem. Soc. 126, 13230-13231 (2004). ;modified after Leskes, Madhu and Vega, Chem. Phys. Lett. to remove center artefact ;using STATES-TPPI ;This pulse program was written according to the corresponding DUMBO-sequence from ;the ENS-Lyon Pulse Program Library ;p9 2.4-4.5 usec, depending on probe deadtime, usually: ;for 200 and 300 MHz, CRAMPS probe required or use 4.5 usec, ;acqu or p9 must be as short as possible, avoiding dipolar coupling effects between DUMBO sequences, ;l11 or d9 must be as large as possible to improve S/N ratio, but keeping acqu positive and small, ;p1 : 90 degree 1H detection pulse ;p2 : presaturation 90 degree pulse ;p9 : acquisition window, 1.7-4.5 usec, depending on probe deadtime ;p10: dumbo-1 pulse for t2 ;p20: dumber-22 pulse for t1 ;p25: = inf1, for t1 increment ;d1 : recycle delay ;d5 : z filter delay, 0.1 μs or multiple of 1/cnst31, otherwise no signal ;d10: parameter for t1 value ;d20: delay between saturation pulses ;l0 : 0 as initial t1 ;l1 : number SPC5 basic cycle elements, for protons 2-4 in real solids ;l3 : t1-increment multiplier, usually 2-4, to reduce required number of rows ;l11: number of oversampled data points to be averaged into one dwell point ;l20: # of pulses in saturation pulse train, 0 if undesired ;pl1 : 1H presaturation power ;pl7 : 1H power for SPC5, B1=5*cnst31 in Hz ;pl12: 1H power for pulses P1 ;pl13: dumbo power ;sp1 : 1H power for windowed dumbo-1 (t2) ;sp2 : 1H power for dumber-22 (t1) (usually somewhat less power than sp1 since ; there is no window), set to pl13 as in setup experiments ;cnst1 : phase for SPC5 reconversion pulse due to t1 evolution period ;cnst31: spinning frequency (usually not more than 15 kHz possible) ;FnMode: undefined ;MC2 : STATES-TPPI ;NS : =16*n ;WDW : F1 QSINE 3, F2 QSINE 2 or EM ;zgoptns :-Dpresat or blank ;$COMMENT=homonuclear decoupling with w-DUMBO ;$CLASS=Solids ;$DIM=2D ;$TYPE=homonuclear decoupling ;$SUBTYPE=explicit acquisition ;$OWNER=hf ;cnst11 : to adjust t=0 for acquisition, if digmod = baseopt "acqt0=1u*cnst11" dwellmode auto #include <Avancesolids.incl> #include <Delayssolids.incl> ; "d3=p9" ;p9 sets the window to make sure it is in microseconds "d9=0.1u*(l11)" ;set the sampling window, defined in Avancesolids.incl "blktr2 = 0.6u" ;this opens the transmitter gate 0.6 usec before the ;pulse, so the transmitter noise is not sampled "l0=0" ;reset F1 dwell counter "inf1=(l3*(2*d3+p20))*2" ;t1 increment "sp1=pl13" "sp2=pl13" define delay dead "dead=1.2u" define delay acqu ;small window, defined by d3, 2.5-4.5 usec depending "acqu=2*p9-1.2u-d9-.1u" ;on probe deadtime ;acqu or p9 must be as short as possible, avoiding dipolar coupling effects ;l11 or d9 must be as large as possible but keeping acqu positive define delay cycle "cycle=4*p9+2*p10+.2u" define loopcounter count "count=aq/cycle" ;make sure td datapoints are sampled define delay rest ;make sure sampling proceeds throughout the sequence "rest=aq-(count*cycle)" define loopcounter count1 ;for STATES-TPPI procedure "count1=td1/2" ;and STATES cos/sin procedure define pulse pul360 "pul360=(1s/cnst31)/5" ;360° pulse define pulse pul90 "pul90=(0.25s/cnst31)/5" ; 90° pulse define pulse pul270 "pul270=(0.75s/cnst31)/5" ;270° pulse "d31=1.0s/cnst31" "p25=inf1" 1 ze ;acquire into a cleared memory "d10=0.1u" ;make sure a short d10 is used initially 2 d31 #ifdef presat ;set with -Dpresat pres, d20 ;delay between saturation pulses (p2 pl1 ph4):f1 ;saturation loop if required lo to pres times l20 #endif /* presat */ d1 ;recycle delay "cnst1=180*cnst31*d10" ;phase correction for SPC5 reconversion pulse, ;due to t1 DQ evolution period, ;defined by the phase-time relationships 10u reset1:f1 ;synchronise pulse and detection RF 1m rpp10 ;reset phase list pointer 1m rpp20 ;reset phase list pointer 1m rpp11 1m rpp12 1m rpp13 1m rpp14 10u pl7:f1 ;SPC5 DQ excitation: 3 pul90:f1 ph11 ipp13 ipp14 ;increment reconversion phase ph13 and ph14 pointers pul360:f1 ph12 ipp12 ;increment phase ph12 pointer pul270:f1 ph11 ipp11 ;increment phase ph11 pointer lo to 3 times l1 5 d3 ;DQ evolution: d3 (p20:sp2 ph20^):f1 ;dumber22 d3 d3 (p20:sp2 ph20^):f1 ;dumber22 lo to 5 times l0 6 pul90:f1 ph13+cnst1 pl7:f1 ;SPC5 DQ reconversion: ;increase ph13 by cnst1 due to evolution period pul360:f1 ph14+cnst1 ipp14 ;increase ph14 by cnst1 due to evolution period ;increment phase ph14 pointer pul270:f1 ph13+cnst1 ipp13 ;increase ph13 by cnst1 due to evolution period ;increment phase ph13 pointer lo to 6 times l1 d5 pl12:f1 ;z filter delay STARTADC ;prepare adc for sampling, set reference frequency, ;defined in Avancesolids.incl RESETPHASE ;reset reference phase (p1 ph1):f1 ;90° detection pulse at pl12 .1u DWL_CLK_ON 7 dead acqu d9 RG_ON .1u RG_OFF ;take l11 complex data points (p10:sp1 ph10^):f1 ;w-dumbo, use 24 usec at 600 MHz or higher dead acqu d9 RG_ON .1u RG_OFF (p10:sp1 ph10^):f1 lo to 7 times count ;make sure td points are sampled rest 1u DWL_CLK_OFF 1m ;DQ filtering (four phase cycling): 1m ip13*16384 ;increments all phases of ph13 by 90° 1m ip14*16384 ;increments all phases of ph14 by 90° rcyc=2 ;next scan ;The rcyc statement is used for acquisition loops ;based on adc rather than go=label. Do not ;specify phase programs behind rcyc 100m wr #0 if #0 zd ;save data 1m ip11*8192 ;increments all phases of ph11 by 45°, ;90° phase for DQ coherence 1m ip12*8192 ;increments all phases of ph12 by 45°, ;90° phase for DQ coherence lo to 2 times 2 ;t1 quadrature detection 8 1m iu0 ;increment counter l0 by 1 lo to 8 times l3 ;for multiple t1 increment "d10=d10+p25" ;p25=inf1=increment for F1 (to make it usec!) ;d10 is the t1 evolution period ;1m rp11 ;reset all phases of ph11, ph12, ph13, and ph14 ;1m rp12 ;to their original values, i.e. to the values they ;1m rp13 ;had before the first ip11, ip12, ip13, and ip14 ;1m rp14 ;in case of STATES remove semicolon at beginning of the 4 lines lo to 2 times count1 ;count1 = td1/2 exit ;finished ph1= 1 1 1 1 2 2 2 2 3 3 3 3 0 0 0 0 ph10= 0 2 ;windowed dumbo phase during t2 ph11= (65536) 0 13107 26214 39322 52429 32768 45875 58982 6554 19661 ph12= (65536) 32768 45875 58982 6554 19661 0 13107 26214 39322 52429 ph13= (65536) 16384 29491 42598 55706 3277 49152 62259 9830 22937 36044 ph14= (65536) 49152 62259 9830 22937 36044 16384 29491 42598 55706 3277 ;an overall constant phase shift of π/2 is applied ;to the reconversion pulse phases ph13 and ph14 for time reversal ph4= 0 ;for presaturation pulse ph20=0 2 ;dumber22 phase during t1 ph30=0 ;needed for acquisition, involved in RESETPHASE ph31=0 2 0 2 1 3 1 3 2 0 2 0 3 1 3 1 ;involved in STARTADC ;ph31 = ph1 + 2*ph13 + 1
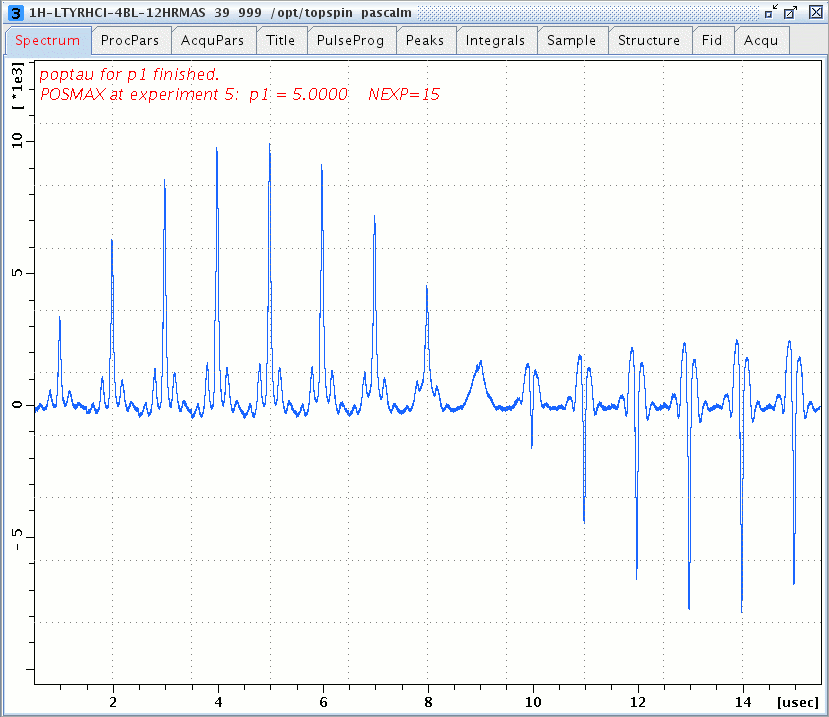
1H MAS spectra of L-Tyrosine.HCl versus the excitation pulse p1 duration (pl1 = 9.8 dB), acquired with a 4-mm diameter, 12-µL HRMAS rotor spinning at 15 kHz, D1 = 5 sec recycle delay, and recorded with Bruker Avance III, 500 MHz WB US magnet.
Pulseprogram parameters for zg:
| General | |
| PULPROG | zg |
| TD | 2048 |
| NS | 4 |
| DS | 0 |
| SWH [Hz] | 100000.00 |
| AQ [s] | 0.0102900 |
| RG | 4 |
| DW [µs] | 5.000 |
| DE [µs] | 6.50 |
| D1 [s] | 5.00000000 |
| TD0 | 1 |
| Channel f1 | |
| NUC1 | 1H |
| P1 [µs] | 5.00 |
| PL1 [dB] | 9.80 |
| PL1W [W] | 33.58852005 |
| SFO1 [MHz] | 500.1666284 |
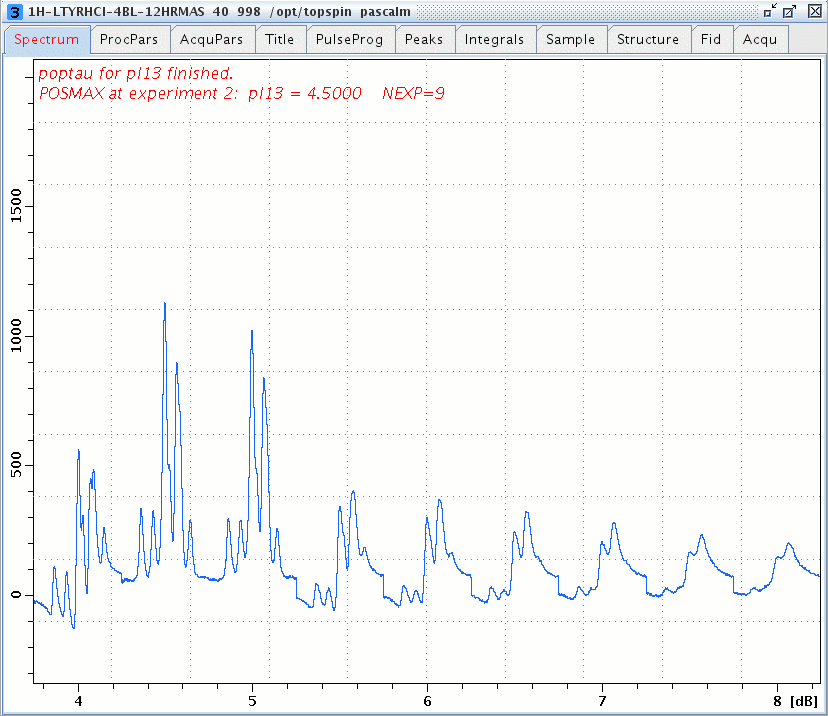
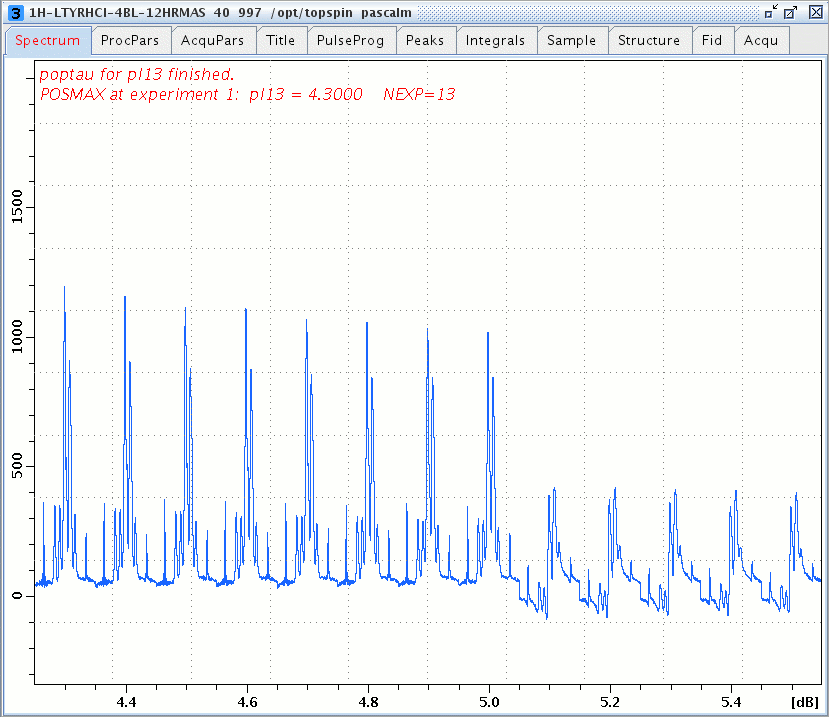
1H windowed DUMBO spectra of L-Tyrosine.HCl versus the DUMBO power pl13, DUMBO pulse duration dumbop = p10 = 24 µsec and rotor spinning at 14 kHz.
Pulseprogram parameters for dumbod2:
| General | |
| PULPROG | dumbod2 |
| TD | 700 |
| NS | 4 |
| DS | 0 |
| SWH [Hz] | 20000.00 |
| AQ [s] | 0.0175750 |
| RG | 4 |
| DW [µs] | 25.000 |
| DE [µs] | 1.10 |
| CNST11 | 0.0000000 |
| D1 [s] | 5.00000000 |
| D3 [s] | 0.00000300 |
| d9 [s] | 0.00000320 |
| L11 | 32 |
| P9 [µs] | 3.00 |
| P10 [µs] | 24.00 |
| PL13 [dB] | 4.30 |
| acqu [s] | 0.00000150 |
| count | 292 |
| cycle [s] | 0.00006010 |
| de [µs] | 0.00 |
| dead [s] | 0.00000120 |
| rest [s] | 0.00000080 |
| Channel f1 | |
| dumbop [µs] | 24.00 |
| NUC1 | 1H |
| P1 [µs] | 5.00 |
| PL1 [dB] | 9.80 |
| PL1W [W] | 33.58852005 |
| PL12 [dB] | 9.80 |
| PL12W [W] | 33.58852005 |
| SFO1 [MHz] | 500.1666284 |
| SP1 [dB] | 4.30 |
| SPNAM1 | dumbo_1+0 |
| SPOAL1 | 0.500 |
| SPOFFS1 [Hz] | 0.00 |
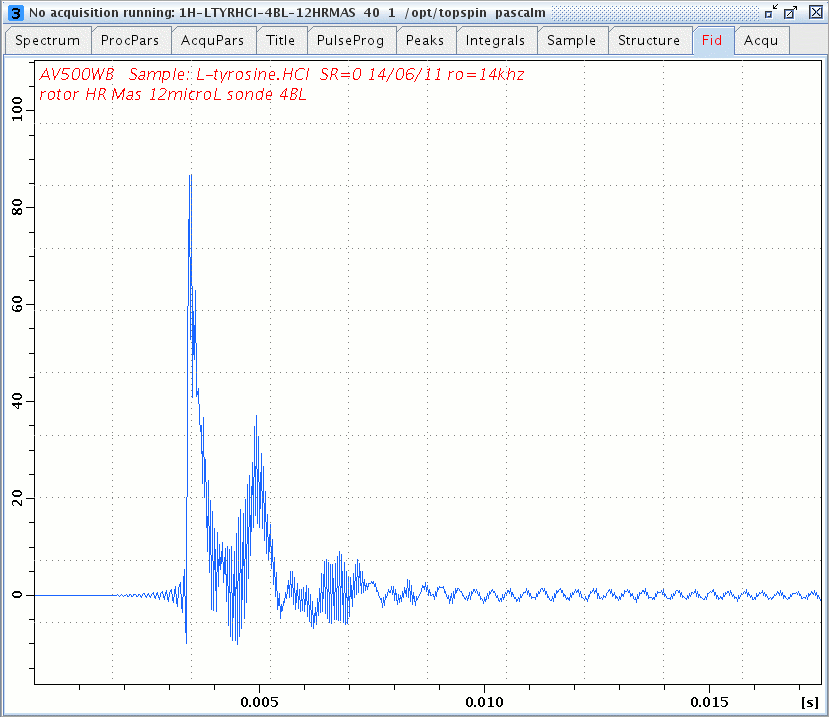
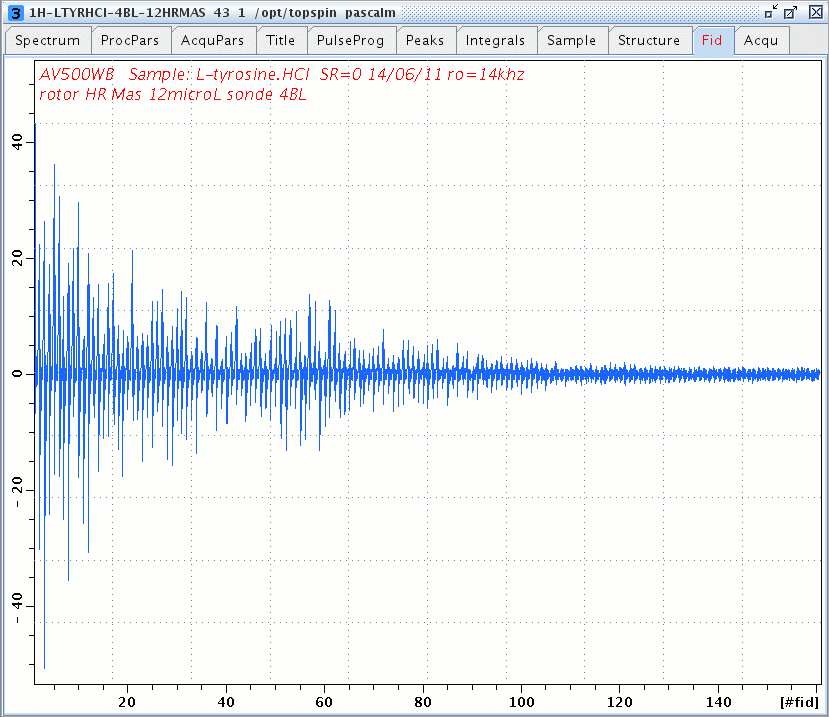
1H windowed DUMBO FID of L-Tyrosine.HCl.
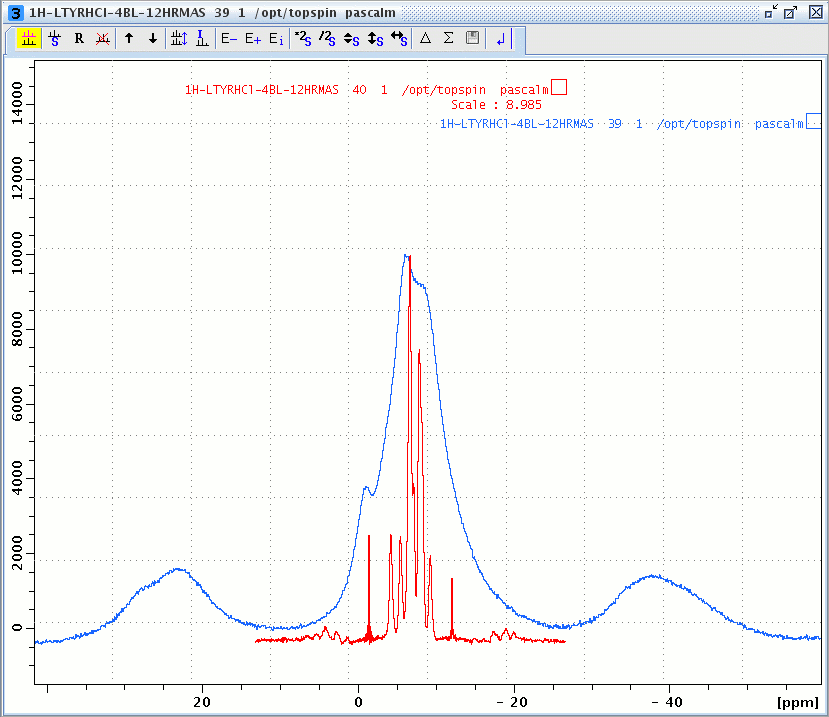
1H windowed DUMBO spectrum (red) and one pulse MAS spectrum (blue) of L-Tyrosine.HCl.
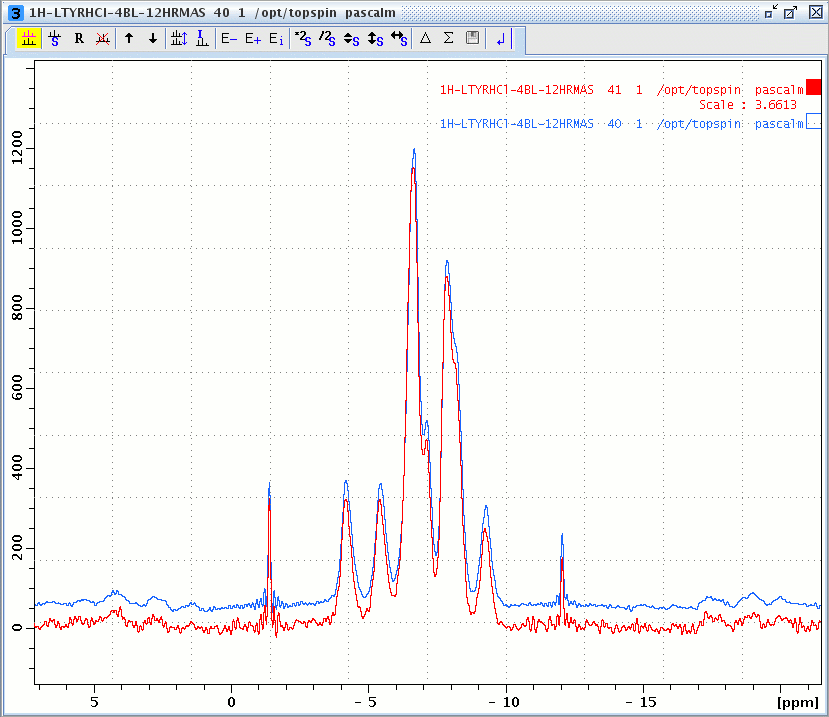
1H windowed DUMBO spectrum of L-Tyrosine.HCl, acquired with the number of oversampling points L11 = 16 (red) and L11 = 32 (blue).
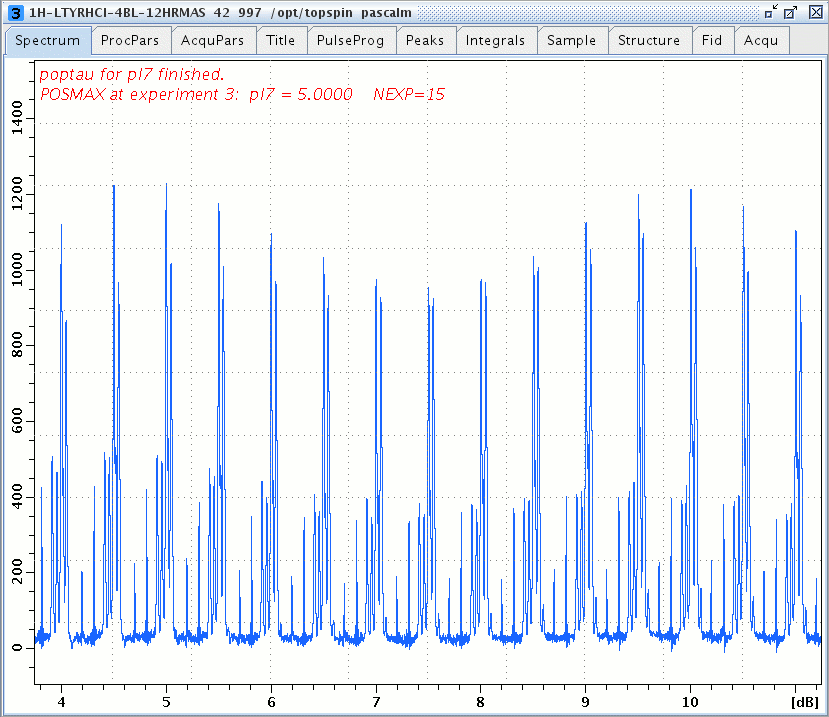
1H DQ/SQ spectrum of L-Tyrosine.HCl versus the SPC5 DQ power pl7, L1 = 4.
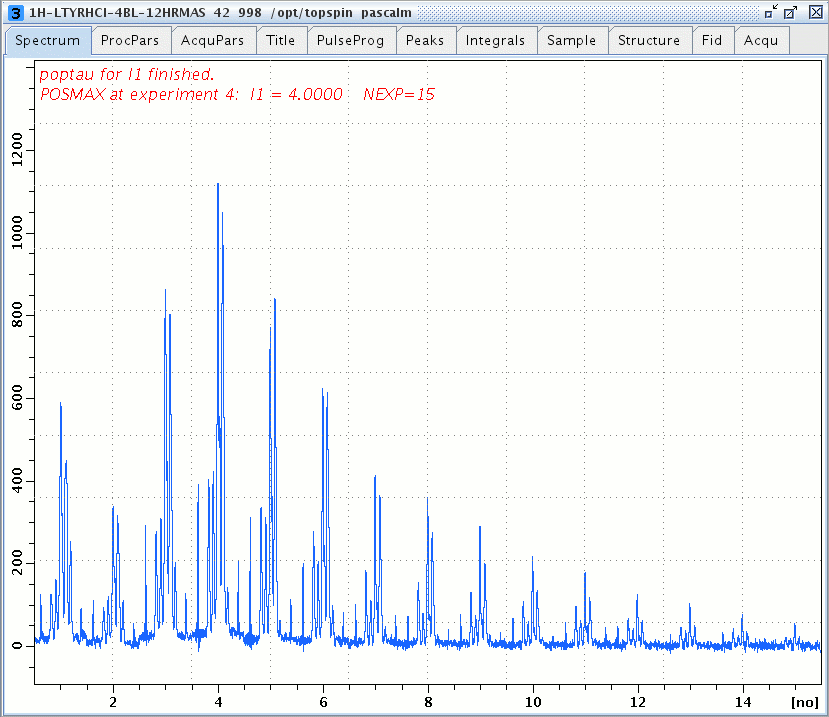
1H DQ/SQ spectrum of L-Tyrosine.HCl versus the number L1 of SPC5 basic cycles, PL7 = 10 dB.
Pulseprogram parameters for DUMBOdqSPC5sqbsw1d.ppm:
| General | |
| PULPROG | DUMBOdqSPC5sqbsw.ppm |
| TD | 700 |
| NS | 16 |
| DS | 0 |
| SWH [Hz] | 20000.00 |
| AQ [s] | 0.0175500 |
| RG | 4 |
| DW [µs] | 25.000 |
| DE [µs] | 1.10 |
| CNST11 | 0.0000000 |
| CNST31 | 14000.0000000 |
| d0 [s] | 0.00000000 |
| D1 [s] | 5.00000000 |
| d3 [s] | 0.00000300 |
| D5 [s] | 0.00000100 |
| d9 [s] | 0.00000320 |
| d31 [s] | 0.00007143 |
| L0 | 0 |
| L1 | 4 |
| L11 | 32 |
| P9 [µs] | 3.00 |
| PL13 [dB] | 4.30 |
| ZGOPTNS | |
| acqu [s] | 0.00000150 |
| count | 291 |
| cycle [s] | 0.00006020 |
| de [µs] | 0.00 |
| dead [s] | 0.00000120 |
| rest [s] | 0.00003180 |
| Channel f1 | |
| CNST1 | 0.000000 |
| NUC1 | 1H |
| P1 [µs] | 5.00 |
| P10 [µs] | 24.00 |
| P20 [µs] | 24.00 |
| PL1 [dB] | 9.80 |
| PL1W [W] | 33.58852005 |
| PL7 [dB] | 10.00 |
| PL7W [W] | 32.07678986 |
| PL12 [dB] | 9.80 |
| PL12W [W] | 33.58852005 |
| pul270 [µs] | 10.71 |
| pul360 [µs] | 14.29 |
| pul90 [µs] | 3.57 |
| SFO1 [MHz] | 500.1666284 |
| SP1 [dB] | 4.30 |
| SP2 [dB] | 4.30 |
| SPNAM1 | dumbo_1+0 |
| SPNAM2 | dumbo_1+0 |
| SPOAL1 | 0.500 |
| SPOAL2 | 0.500 |
| SPOFFS1 [Hz] | 0.00 |
| SPOFFS2 [Hz] | 0.00 |
The duration of the basic SPC5 cycle is pul270 + pul360 + pul90 = 28.57 µsec. Two basic cycles last 57.14 µsec.
In the evolution period, the duration due to the two DUMBO pulses and the four d3 delays is 2*p20 + 4*d3 = 60 µsec. Since there is no signal sampling during this period, we can decrease the duration of d3 from 3 down to 2.285 µsec so that inf1 = p25 = 57.14 µsec. The 2D spectrum will be on-resonance in the F1 dimension if it is on-resonance in the F2 dimension.
1H DQ/SQ 2D ser file of L-Tyrosine.HCl, showing the number of FID for the F1 dimension.
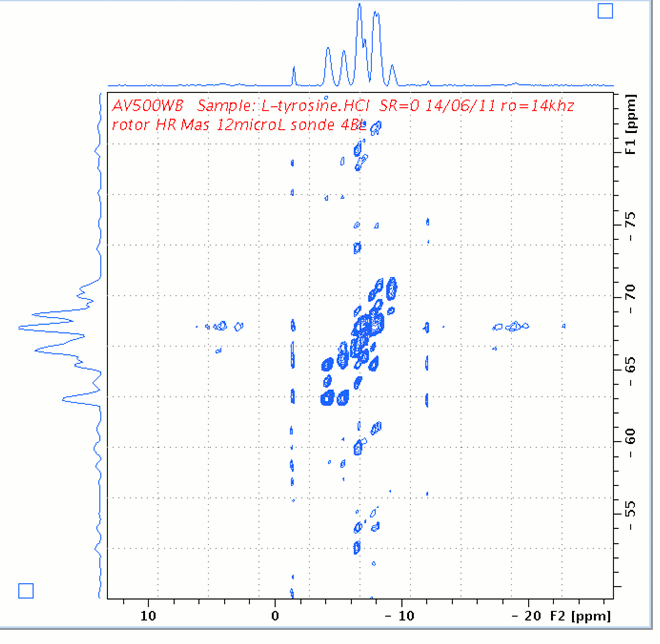
1H DQ/SQ 2D spectrum of L-Tyrosine.HCl.
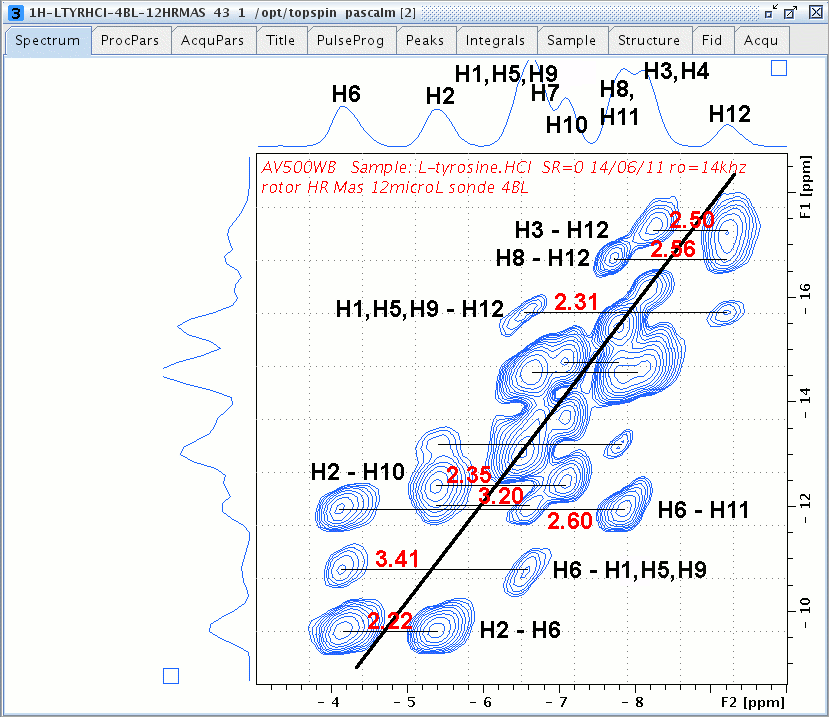
Zoomed 1H DQ/SQ 2D spectrum of L-Tyrosine.HCl, red-coloured numbers are distances separating two protons in Å unit from Luis Mafra et coworkers, J. Magn. Reson. 199, 111-114 (2009).
Pulseprogram parameters for DUMBOdqSPC5sqbsw.ppm:
| General | |
| PULPROG | DUMBOdqSPC5sqbsw.ppm |
| TD | 700 |
| NS | 16 |
| DS | 0 |
| SWH [Hz] | 20000.00 |
| AQ [s] | 0.0175500 |
| RG | 4 |
| DW [µs] | 25.000 |
| DE [µs] | 1.10 |
| CNST11 | 0.0000000 |
| CNST31 | 14000.0000000 |
| D1 [s] | 5.00000000 |
| d3 [s] | 0.00000228 |
| D5 [s] | 0.00000100 |
| d9 [s] | 0.00000320 |
| D10 [s] | 0.00000010 |
| d31 [s] | 0.00007143 |
| inf1 [µs] | 57.14 |
| L0 | 0 |
| L1 | 4 |
| L3 | 1 |
| L11 | 32 |
| P9 [µs] | 3.00 |
| p25 [µs] | 57.14 |
| PL13 [dB] | 4.30 |
| ZGOPTNS | |
| acqu [s] | 0.00000150 |
| count | 291 |
| count1 | 80 |
| cycle [s] | 0.00006020 |
| de [µs] | 0.00 |
| dead [s] | 0.00000120 |
| rest [s] | 0.00003180 |
| Channel f1 | |
| CNST1 | 1.0000000 |
| NUC1 | 1H |
| P1 [µs] | 5.00 |
| P10 [µs] | 24.00 |
| P20 [µs] | 24.00 |
| PL1 [dB] | 9.80 |
| PL1W [W] | 33.58852005 |
| PL7 [dB] | 10.00 |
| PL7W [W] | 32.07678986 |
| PL12 [dB] | 9.80 |
| PL12W [W] | 33.58852005 |
| pul270 [µs] | 10.71 |
| pul360 [µs] | 14.29 |
| pul90 [µs] | 3.57 |
| SFO1 [MHz] | 500.1666284 |
| SP1 [dB] | 4.30 |
| SP2 [dB] | 4.30 |
| SPNAM1 | dumbo_1+0 |
| SPNAM2 | dumbo_1+0 |
| SPOAL1 | 0.500 |
| SPOAL2 | 0.500 |
| SPOFFS1 [Hz] | 0.00 |
| SPOFFS2 [Hz] | 0.00 |
Acquisition parameters:
| F2 | F1 | |
| Experiment | ||
| PULPROG | DUMBOdqSPC5sqbsw.ppm | |
| AQ_mod | DQD | |
| FnMODE | undefined | |
| TD | 700 | 160 |
| NS | 16 | |
| DS | 0 | |
| TD0 | 1 | |
| Width | ||
| SW [ppm] | 39.9866 | 34.9901 |
| SWH [Hz] | 20000.00 | 17500.875 |
| IN_F [µs] | 57.14 | |
| AQ [s] | 0.0175500 | 0.0045712 |
| Nucleus1 | ||
| NUC1 | 1H | 1H |
| O1 [Hz] | -3371.57 | -3371.57 |
| O1P [ppm] | -6.741 | -6.741 |
| SFO1 [MHz] | 500.1666284 | 500.1666284 |
| BF1 [MHz] | 500.1700000 | 500.1700000 |
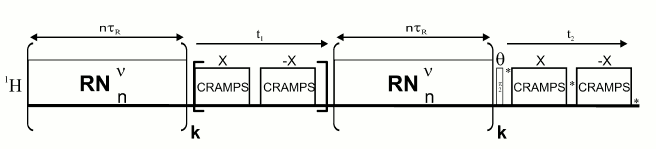
RN-DQ/SQ-DUMBO excitation pulse sequence.
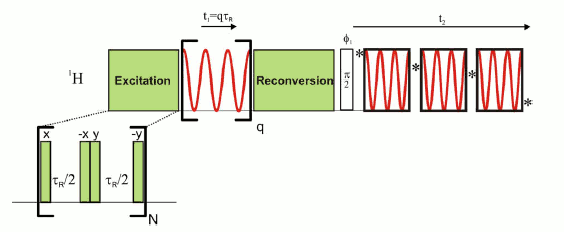
BABA-DQ/SQ-SAM excitation pulse sequence.
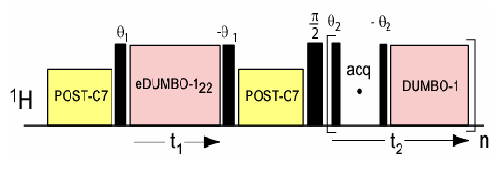
DQ-DUMBO excitation pulse sequence.