*** Outline ***
Denoising an FID with SVD
Denoising an FID increases the signal to noise ratio of the corresponding spectrum without broadening the linewidth.
2012 SVD Java application
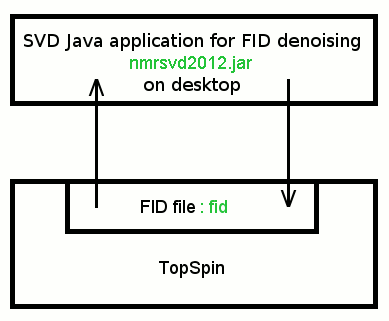
We provide a new Java application (nmrsvd2012.jar), which allows us to denoise FID signals acquired with Bruker Avance spectrometer. SVD Java application and TopSpin exchange FID data via TopSpin 4-octet integer binary file: fid.
TopSpin FID file is a 4-octet little-endian integer binary file. On the other hand Java integer number is a 4-octet big-endian number. File conversion is required.
Installation
Download the Java 1.4 application nmrsvd2012.jar (last updated March 26, 2012) on the desktop of the computer. It consists of two views: matrix and SVD.
Execution
In TopSpin:
- Make a copy of experiment data with the wrpa command then display the FID.
- Apply the convdta command if data are obtained in digital mode.
On desktop:
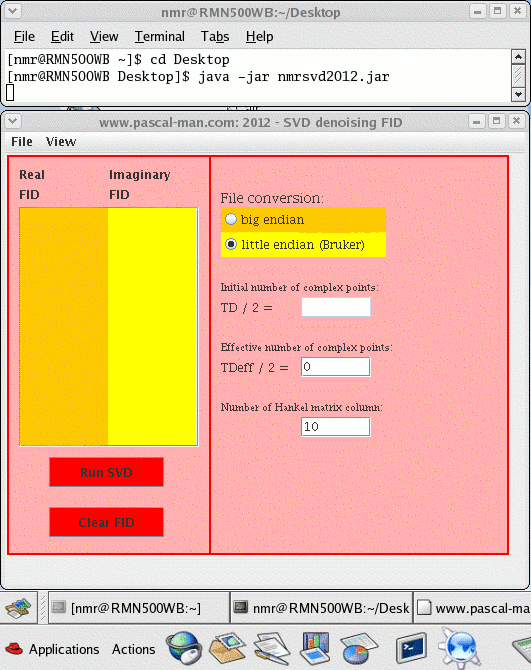
- Double click the nmrsvd2012.jar icon to start the program. The matrix view appears.
- If necessary, check big-endian radio-button for file conversion of other spectrometers. Default state is little-endian radio-button for Bruker TopSpin checked.
- Read the noisy FID binary file fid via File / Open 4-octet integer binary file... menu item, the data are displayed in the two textArea as integer values.
- Make a copy of the original noisy FID binary file (fid) via File / Save original 4-octet integer binary file... menu item, the saved data are those (integer values) displayed in the two textArea.
- Provide the number of complex points for denoising: TDeff/2 smaller or equal to TD/2.
- Provide the number of Hankel matrix columns, it must be smaller or equal to 0.5*TDeff/2.
- Click Run SVD button for denoising. The duration of calculation depends on the number of Hankel matrix columns and the size of random access memory. The singular values and their graph are shown in the SVD view.
- Save this lengthy decomposition on hard disk file using File / Save SVD... menu item in the SVD view.
- Provide a reduced number of singular values in the white textField (this number should be smaller than the number of Hankel matrix columns), then click Hankel button. The denoised FID (non-integer values) is shown in the two textArea of the matrix view.
- Replace the noisy FID binary file fid with denoised data by selecting File / Save denoised fid as binary... menu item.
In TopSpin:
- Display the denoised FID file in TopSpin, then process the data.
- Jump to Point 7 of "On desktop" part for more denoising if the signal to noise ratio is not good enough. Or recall the saved SV decompostion in Point 8 of "On desktop" part using File / Open SVD... menu item in the SVD view; then jump to Point 9.
Avance spectrometer with Linux operating system
To start nmrsvd2012.jar in Linux OS, first launch a terminal window (Desktop - Shell - Konsole) called nmr@RMN500WB in our case.
(1) Check the version of Java: java -version
(2) Find the location of Desktop : ls
(3) Move to Desktop directory: cd Desktop
(4) Launch nmrsvd2012.jar: java -jar nmrsvd2012.jar